Summary
In late December 2019, a new disease named COVID-19 (COronaVirus Infectious Disease 2019), due to a new Betacoronavirus, the SARS-CoV-2, was notified in Wuhan, China. WHO declares “A public health emergency of international concern” on January 30th, 2020. A few weeks later, the virus has spread in more than 170 countries and has caused more than 215,000 cases including more than 8,790 deaths (with a large majority in China – 18th March 2020, WHO).
Preliminary investigations on available samples collected in 2013 gave a mean nucleotide identity of 96% with the genome of a Sarbecovirus (genus Betacoronavirus) of a Rhinolophidae bat specimen. Despite these announcements, the most recent and direct ancestor of this emergent virus is yet to be discovered and the natural history of its emergence remains to be elucidated. This brings both the WHO and the French National Agency for Research (ANR) to list the quest for the reservoir and the natural history of the emergence as a top priority. DisCoVER aims at answering the question of the origin of SARS-CoV by bringing together an interdisciplinary team of experts in the field from the Caen University, IRD, CNRS, Kasetsart University, Mahidol University and Center of Infectiology Lao Christophe Mérieux.
The main objective of the project is to track the origin of SARS-CoV2 in natural settings sharing biogeographical and socio-ecological features with South-Western China (northern regions of Thailand and Lao PDR). The aim is to characterise the SARS-CoV2 natural cycle and the modalities of its emergence in humans. The zoonotic/emergence risk of SARS-CoV2-related Sarbecovirus members infecting wild animals in northern Southeast Asia will be estimated using a model that will integrate phylodynamic data/analyses with socio-ecological factors to develop real strategies for anticipation and prevention of future emergence.
Proposal description
Scientific and societal context
In late December 2019, a new disease named COVID-19 (COronaVirus Infectious Disease 2019), due to a new Betacoronavirus, the SARS-CoV-2, was notified in Wuhan, China. WHO declared “A public health emergency of international concern” on January 30th, 2020. Six weeks later, the virus has spread to more than 170 countries and has caused more than 215,000 cases including more than 8,790 deaths (with a large majority in China – 18th March 2020, WHO). The first investigations undertaken to identify the origin of this epidemic showed a mean nucleotide identity of 96% with the genome of a Betacoronavirus (one of the four genera in the Coronaviridae family) of a bat specimen (Rhinolophus affinis) captured in South-Western China (Yunnan province) by collaborators in 2013 (Zhou,[…] Shi et al. 2020). Moreover, a pangolin coronavirus is suspected to have partially contributed to the genesis of SARS-CoV2 by recombination (affecting a very minor part of the receptor binding domain, key for human infection) (Xiao, et al. 2020). Despite these announcements, the most recent and direct ancestor of this emergent virus is yet to be discovered and the natural history of its emergence remains to be elucidated. This brings both the WHO and the French National Agency for Research (ANR) to list the quest for the reservoir and the natural history of the emergence as a top priority. DisCOVER is designed toward the investigation of these aspects.
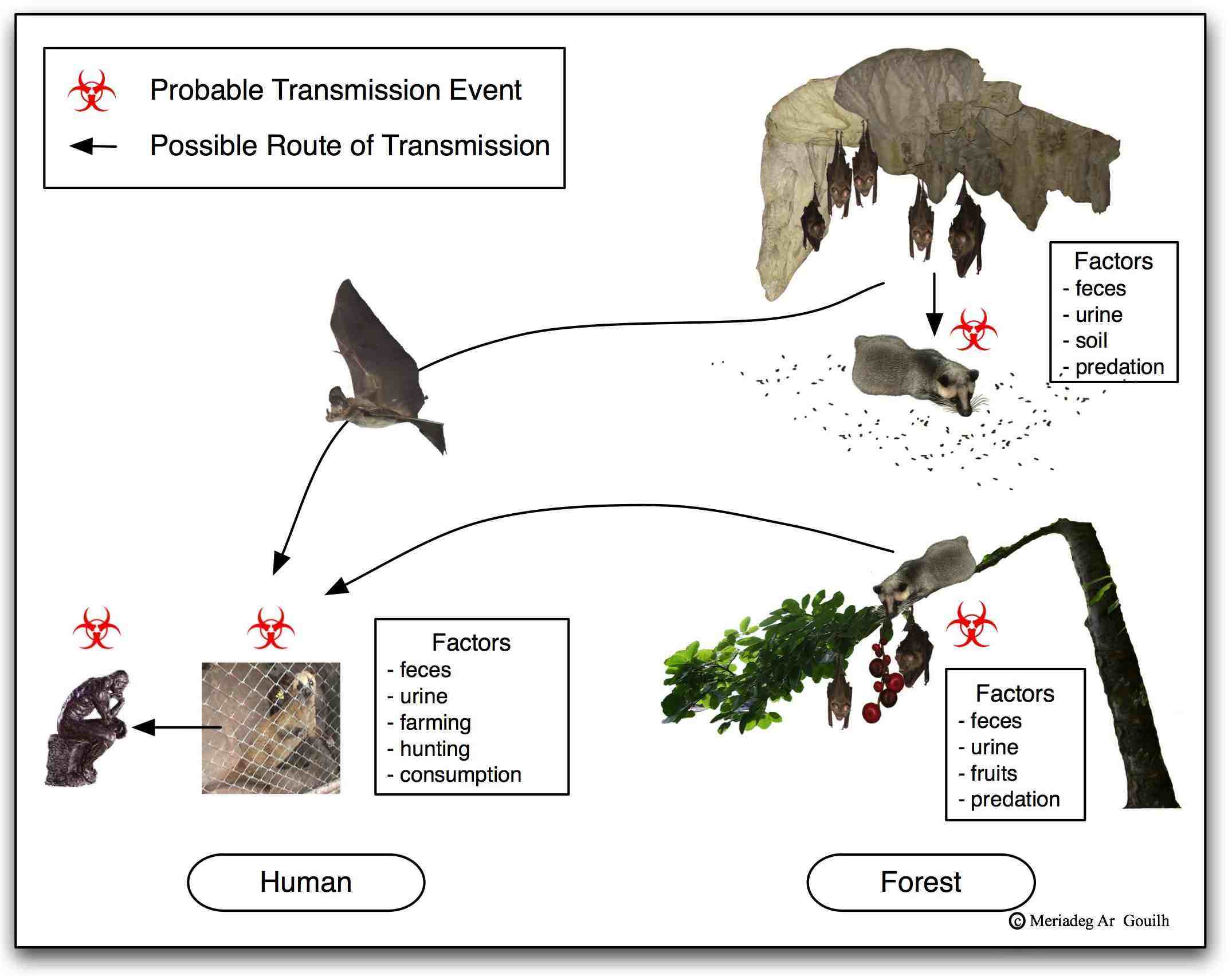
Epidemiological and virological data point out a unique origin (or restricted in time and space) of human emergence around a wet (living animals) market in the city of Wuhan, without precise information released (notably on the type of samples tested). By analogy with the emergence of SARS-CoV (2002-2003; Cf. State of the Art section), and other coronaviruses natural history, the existence of an intermediate host, perhaps sold on the Wuhan market, seems likely to have initiated such an epidemic. The diversity of wild fauna (notably in southern China and in South-East Asia, where bat diversity peaks to over a hundred species), associated with coronaviruses wide circulation, evolutionary and zoonotic potential, indicate that one may expect other diseases emergence where socio-ecological characteristics are similar to those of South-Western China.
The identification of the reservoir and intermediate hosts species, the description of the viral diversity in wild species (with particular focus on Rhinolophidae) and the precise characterisation of the chain of events leading to human infection therefore seems essential for: i) on one hand, urgently implement appropriate measures to avoid new contaminations from animal sources, in China and in neighbouring countries (in the current epidemic context where attention is drawn on a specific virus), and ii) on the other hand, develop real strategies for anticipation and prevention of future emergences.
The main objective of the project is to track the origin of SARS-CoV2 and to characterise its natural cycle, the modalities of its emergence in humans, and assess the zoonotic/emergence risk of other Betacoronavirus members infecting wild animals in Southeast Asia. Data will be used to target geographical zones and localities in which risk estimation would allow us to provide basic guidelines at destination of stakeholders in order for them to implement appropriate risk mitigation measures.
Specific objectives are to:
1) (WP2) Estimate prevalence and diversity of SARS-CoV2 and related betacoronaviruses in the study area.
2) (WP3) Assess the natural cycle of SARS-CoV2 and related viruses in wildlife by exploring:
a) temporal evolution of excretion in reservoir species (eco-physiological & environmental factors)
b) inter-species spillover (differentiating reservoir and intermediate hosts)
c) the evolution of the SARS-CoV2 clade using a time-resolved / ecology-informed phylogeography
3) (WP4) Estimate the zoonotic risk and human emergence potential of Betacoronavirus / Sarbecovirus clade members. We will build a model that will integrate all previously mentioned data/analyses with socio-ecological factors (human population densities and movements, market wildlife encroachment, land-use, farming practices, wildlife trade etc…).
State of the art
CORONAVIRUSES – A great number of both mammalian and avian species composes the wide host spectrum of coronaviruses. They are responsible for essentially respiratory and enteric pathologies with variable severity. The IBV or “infectious bronchitis virus” in domestic fowl, the TGEV “transmissible gastroenteritis coronavirus” in domestic pigs and the MHV or “mouse hepatitis virus” were the first animal coronaviruses described in the 1930s. These viruses were first studied in veterinary science because they were responsible for severe pathologies in their host species. Human coronaviruses OC43 and 229E were described in the 1960s, but have attracted limited interest because of the mild respiratory symptomatology which they were linked to.
OC43 and 229E emerged in human population at least one to two centuries ago and resulted from a spillover (i.e. interspecies transmission) from bovine coronavirus and a coronavirus harboured by bats, respectively. Bats are known to harbour an astonishing diversity of coronaviruses (Ar Gouilh et al. 2018) among which Betacoronavirus, and more precisely the subgenus Sarbecovirus, accounts for 4 major clades described to date: i) the very diversified clade from which emerged SARS-CoV, ii) its sister-group, from which originates the SARS-CoV2, the Western Palearctic clade divided in iii) the European clade (for which the evolution is under close monitoring for 6 years, by the coordinator of this proposal thanks to the EPICOREM ANR funded project) and the African clade which is yet poorly documented.
EMERGENCE: SARS-CoV, MERS-CoV and SARS-CoV2 – The evolutive and emerging potential of coronaviruses came into the limelight during the 2002-2003 SARS (Severe Acute Respiratory Syndrome) pandemic with over 8,000 cases including 774 deaths (http://www.who.int/csr/sars/en/). The origin of SARS-CoV has been traced back to Rhinolophidae bat species but its emergence in Human is undoubtedly linked to wildlife hunting, farming and trading of civet (Paguma larvata, identified as the intermediary-host) in the Guangdong province (Ge et al. 2013). This South-Western China region experienced an outstanding industrialisation, urbanisation and growth during decades preceding the SARS-CoV emergence. The number of civet farming facilities expanded fifty times during 5 years before the emergence, illustrating te consequences of uncontrolled human activities at the interface with wildlife. Another similar and prominent spillover of coronavirus occurred in the region in 2016 and provoked a large-scale outbreak of fatal disease in pigs (SADS-CoV), causing the death of over 24,000 piglets in South-Western China (Zhou et al. 2018). The event of the SARS pandemic claims several features that are unique to it: it was the first of the 21st century pandemics. It was the first pandemic to benefit from Internet networking along with tight international collaboration between research laboratories, and the first to be brought under control thanks to the rapid launching of sound health policy. Following the SARS pandemic, two other human coronaviruses (NL63 and HKU1, both of animal origin) were identified albeit infecting humans for decades or centuries.
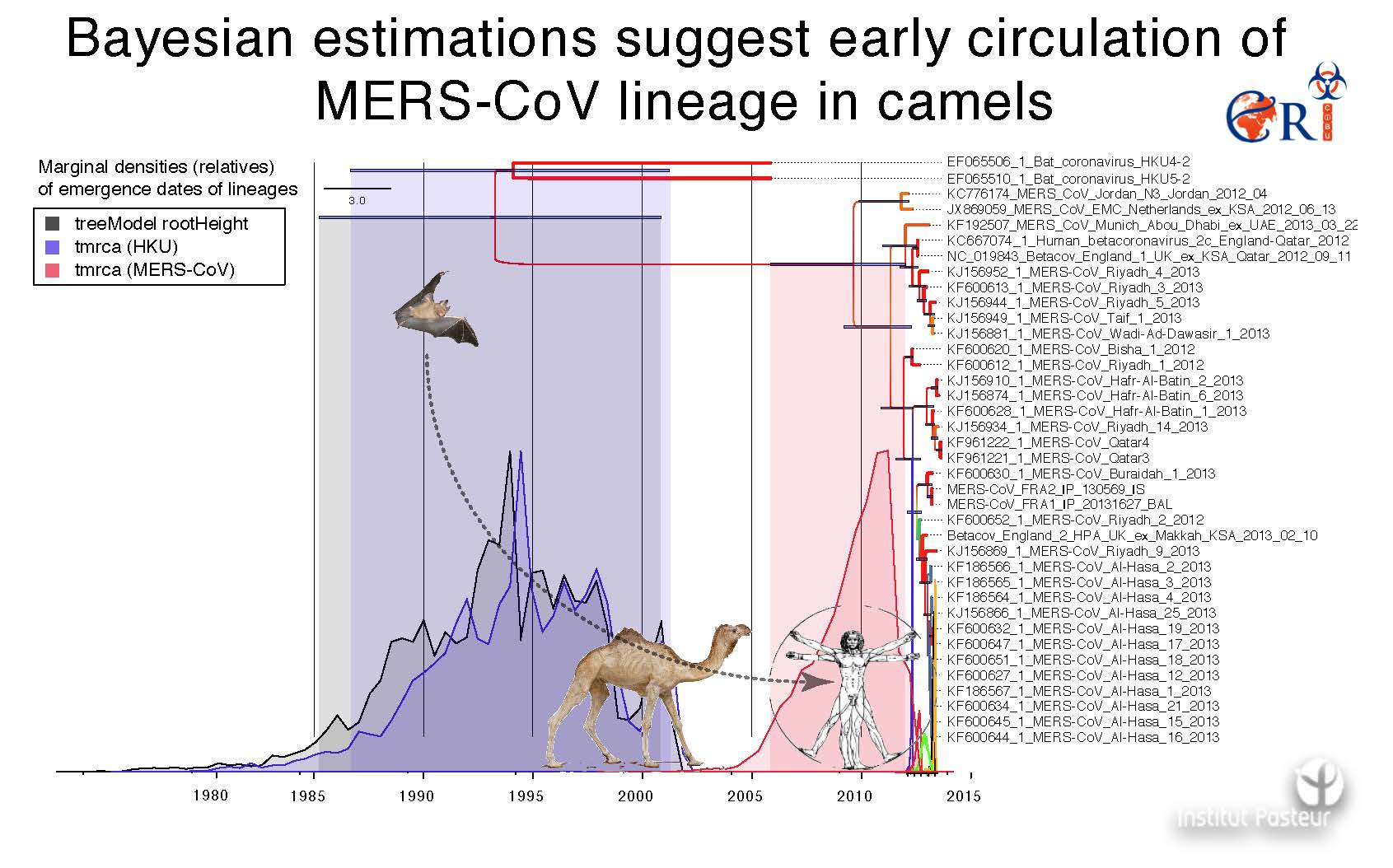
Ten years later, in April 2012, another Betacoronavirus, the Middle-East Respiratory Syndrome Coronavirus (MERS-CoV), emerged and was responsible for a SARS-like syndrome and acute renal failure in humans. To date, a total of 2,519 lab-confirmed cases are reported, including 866 (37%) associated deaths (WHO, MERS situation update, Jan 2020). This coronavirus found his deep origins in bats but circulated in camels for over 40 years. Camel extraordinary abundance in the vicinity of human in the middle east promoted the species barrier crossing and the emergence of MERS-CoV in human, yet without very efficient inter-human transmission in the global population.
These examples of emergence, extensively studied, revealed that the probability of spillover (i.e. interspecies transmission) of a virus is all the more important as the contacts are frequent. This makes the “farming” of a wild animal (susceptible to virus infection) particularly at risk for urban and immunologically naive human populations. The actual SARS-CoV2 emergence and pandemic are a reminder of the permanent risk of emergence of zoonotic pathogens spreading from their animal reservoirs to humans, and the need for better understanding of underlying processes as a first step toward the anticipation and the prevention of re-emergence. The economic cost of the SARS pandemic was estimated to be between 30 and 50 billions USD, though the virus circulated only 8 months in the human population and infected less than 9,000 individuals on the planet. Given China, USA, or European Commission recent announcements, SARS-CoV2 pandemic is going to overcome SARS-CoV in term of global cost. A deeper understanding of the transmission ecology of coronaviruses and zoonoses, especially at the interface between humans and animals, is necessary to improve anticipation-based prevention measures to mitigate the risk of emergence.
Such vision is an integral part of the “One Health” approach that incorporates broader, more holistic perspective to the continuum among ecosystems. This approach allows us using tools of preventive action, namely rapid detection of a sporadic yet “abnormal” phenomenon and the implementation of a rapid and adequate response.
Most studies on SARS-CoV2 and funding engaged to date are understandably directed toward emergency measures such as clinical management of cases, potential drug trials or vaccine development with little to no attention paid to the necessity to increase the knowledge on the diversity and origin of the virus. Data from China on this crucial question have been eclipsed, in a way or another, by the actual human crisis. It is our firm conviction that these aspects need urgent and adequate consideration and should be explored shortly. Indeed, given the fast evolution rate of the coronavirus(es) we are dealing with, the footprints of its emergence is going to rapidly be blurred and need to be immediately and properly investigated, to document our future emergences anticipation and prevention program. To our knowledge, no other consortium had the perfect mixture of advantages and prompt in-situ feasibility proposed in DisCoVer. This project aims to pave the way for future programs dedicated to anticipating emerging infections. As told by Charles Nicolle, director of the Institut Pasteur de Tunis, one century ago: “Il y aura des maladies nouvelles, c’est un fait fatal / there will be new diseases, it is an inevitable fact”. This is now our duty to address his visionary legacy.


Share this Post
